Checking Multivariate Normality in a Lavaan Model
(SEM, CFA, Path Analysis)
by Arndt Regorz, MSc.
July 07, 2024
As a default a lavaan model is estimated using a maximum likelihood (ML) estimator. A crucial assumption of the ML estimation is multivariate normality. This blog post shows you how to check the normality assumption for a structural equation model (SEM), a confirmatory factor analysis (CFA) or a path analysis with R.
1. What Exactly is the Multivariate Normality Assumption?
The normality assumption does not apply to all variables in the model, only to endogenous observed variables, that is, observed variables that are predicted by other variables (observed or latent) in the model.
For an SEM or CFA these are the indicators of latent variables (factors), because in a reflective measurement model these are conceptualized to be influenced by the latent factors. If your model contains measured covariates the normality assumption does not extend to them as long as they are exogenous (e.g., age as a covariate).
In a standard path analysis observed endogenous variables are the dependent variables and mediators, not the independent variables.
But what happens if a binary exogenous variable is part of the model (e.g., gender)? In that case the assumption is conditional multivariate normality, i.e. conditional on the value of the binary predictor variable (or in the case of more than one binary exogenous variable conditional on the combination of the binary exogenous variables).
2. How to Test Normality in an SEM or a CFA?
First, you have to filter out all variables from your dataframe that are not indicator variables in your CFA or SEM (for the following code example saving it in the dataframe data_indicators). Then you can run Mardia’s test on the resulting dataframe, using the MVN package.
library(MVN)
mvn(data_indicators, mvnTest="mardia", univariateTest = "SW")
Figure 1
Testing Multivariate Normality
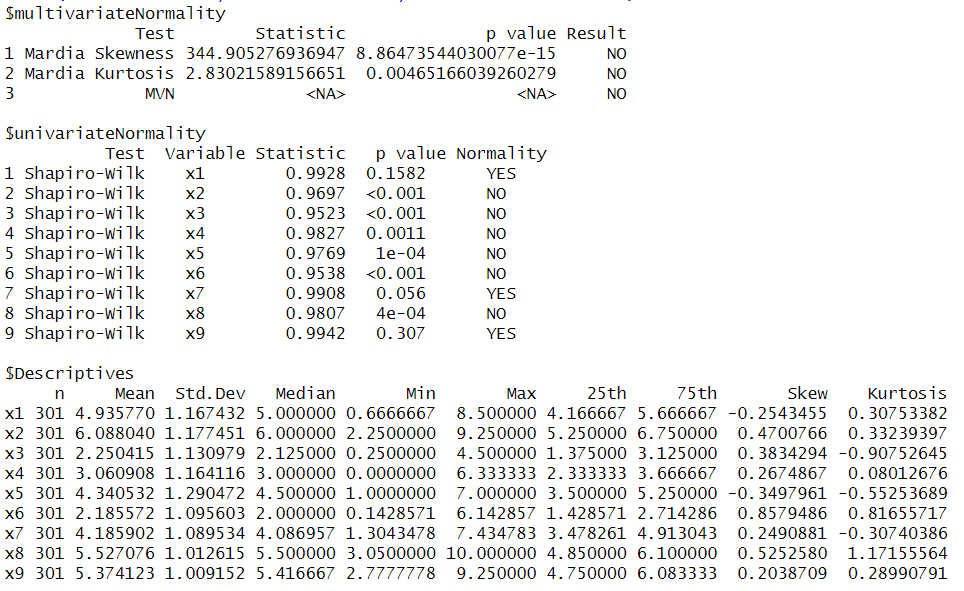
There are three blocks in the output: Multivariate normality, univariate normality, and descriptives. The key results are to be found in the first block.
1 Multivariate skewness: The test is significant, indicating a skewness that is not consistent with multivariate normality - Result NO.
2 Multivariate kurtosis: The test is significant, indicating a kurtosis that is not consistent with multivariate normality - Result NO.
3 MVN (multivariate normality): If at least one of the two tests above is significant then there is no multivariate normality - Result NO.
3 MVN (multivariate normality): If at least one of the two tests above is significant then there is no multivariate normality - Result NO.
If you want to know which variables contribute to the nonnormality you can find univariate normality tests (Shapiro-Wilk) in the second block of the output.
In the third block of the output you can find columns for (univariate) skewness and kurtosis.
3. How to Test Normality in a SEM or CFA with a Binary Covariate
In the presence of a binary covariate the assumption changes to conditional multivariate normality. Therefore, you have first to split the dataframe by the binary covariate, in the following example by sex:
library(dplyr)
data_male <- filter(data_full, sex = 0)
data_female <- filter(data_full, sex = 1)
Then you run Mardia’s test separately in both new dataframes.
4. How to Test Normality in a Path Analysis
Since only the endogenous variables have to be normally distributed the multivariate normality test is run only for those variables, i.e. mediators and dependent variables.
First you have to filter out the exogenous variables from the dataset (in the following example: MED1, MED2, MED3, DV1, DV2).
library(dplyr)
data_endo <- select(data_full, MED1, MED2, MED3, DV1, DV2)
Then, you run the multivariate normality test.
library(MVN)
mvn(data_endo, mvnTest="mardia", univariateTest = "SW")
If one of your independent variables is a binary variable you have to test for conditional multivariate normality in a simular way to the preceding section of this blog post.
5. What to do if the Normality Assumption is Violated?
Lavaan provides a number of robust estimators that do not require normality in order to get correct results.
One of the most popular estimators is estimation with a Satorra-Bentler correction, to be included in the sem() or cfa() function:
estimator="MLM"
Unfortunately, the Satorra-Bentler estimator works only with complete data. Therefore, if you use full information maximum likelihood (FIML) to estimate a model with missing data you need a different robust estimator. Here, the Huber-White estimator (also known as sandwich estimator) can be deployed:
missing = "FIML", estimator="MLR"
Another available robust estimation option is bootstrapping.
References
Brown, T. A. (2015). Confirmatory factor analysis for applied research. Guilford publications.
Kline, R. B. (2023). Assumptions in structural equation modeling. In R. H. Hoyle (Ed.), Handbook of structural equation modeling (2nd ed., pp. 128-144). Guildford Press.
Citation
Regorz, A. (2024, July 07). Checking multivariate normality in a lavaan model (SEM, CFA, path analysis). Regorz Statistik. https://www.regorz-statistik.de/blog/lavaan_normality.html
Other Blog Posts You Might Find Interesting
SEM/CFA: Checking the Linearity Assumption in R/lavaan